
Hello,
This newsletter features some updates made to The Triticeae Toolbox (T3) over the last few months:
- Changes we've made to reduce the number of duplicate accessions in the database. In this case, duplicate accessions are the same germplasm but have multiple entries in the database due to variations in spelling or punctuation or use a synonym.
- Changes to the accession upload process that make it easier to add pedigrees while uploading new accessions.
- Improvements to the custom barcode label designer. These changes allow the user to create plot-level barcodes for multiple trials at once and have more control over the order of the generated labels.
- We also want to highlight our ongoing collaboration with GrainGenes, a USDA-ARS database hosting peer-reviewed genomic data.
Reducing Duplicate Accessions
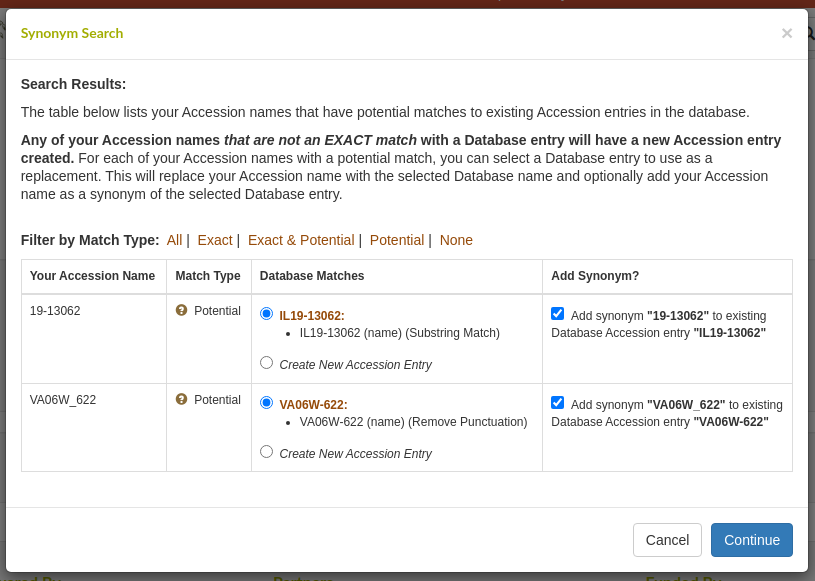
When uploading new accessions, it is important to avoid creating duplicates that differ from existing accessions just by punctuation or letter-case. To facilitate identifying such duplicates, T3 has created a standalone synonym search tool, available at https://synonyms.triticeaetoolbox.org. You can use this tool to paste in a list of germplasm names and the tool will search the selected database and try to find matching accession entries - even if the name or synonym varies in letter case, spelling, or punctuation. The user can then mark any of the results as a match and download a file containing all of the matches. However, this requires the user to manually replace any matches in an accession or trial upload template before uploading it to T3.
Now, the synonym search tool has been integrated into the accession and trial upload workflows on T3. When a template is uploaded to T3, all of the accession names used in the template will first be sent to the synonym search tool and any potential matches to existing accession entries will be displayed to the user before the template is processed. This allows the user to select an existing accession entry as a match and the name in the template will be automatically replaced with the existing name before the file is saved.
Accession Upload Improvements
The accession upload template has been simplified so only the columns of properties you are adding/updating need to be included. Now, the only two columns that are required are the accession_name and species_name. All other columns are optional and can be included in any order.
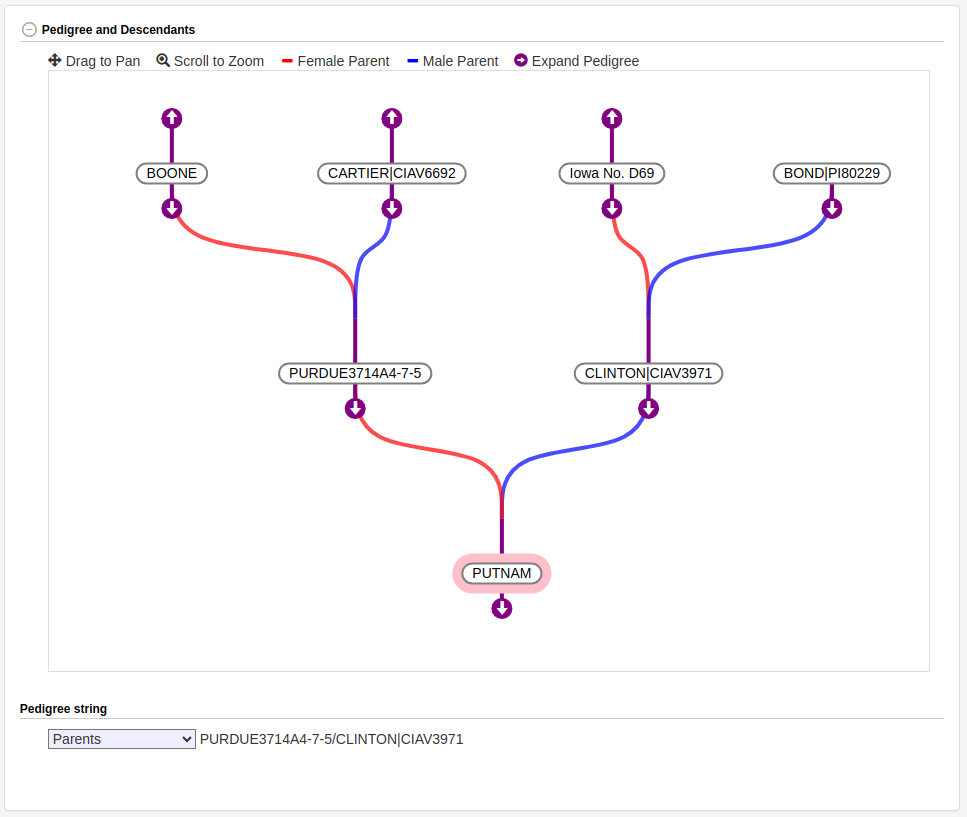
There are also new columns that can be used to add parsed pedigrees. These are different than the existing purdy pedigree string, which is a free text property that gets displayed exactly as it's stored. The parsed pedigrees are direct links between a progeny accession entry and accession entries for its parents. To add a parsed pedigree for an accession, add the names of the parents to the female_parent and male_parent columns. If an accession entry exists in the database with the same name, the progeny will be linked to that existing entry. Any missing parents will be automatically created for you. Once the parent links are established, the pedigree can be viewed in the interactive pedigree viewer - which you can use to browse up and down a pedigree tree.
In addition, most of the uploads have been updated to include support for the newer .xlsx Excel format. This includes the accession upload and trial upload.
Label Designer Improvements
The label designer is a tool that allows you to create your own barcodes that can be printed onto label sticker paper. The barcodes can then later be scanned when you want to reliably and quickly identify samples. You can generate labels for accessions, seedlots, plots within a field trial, wells of a genotyping plate and anything else you can make a list of.
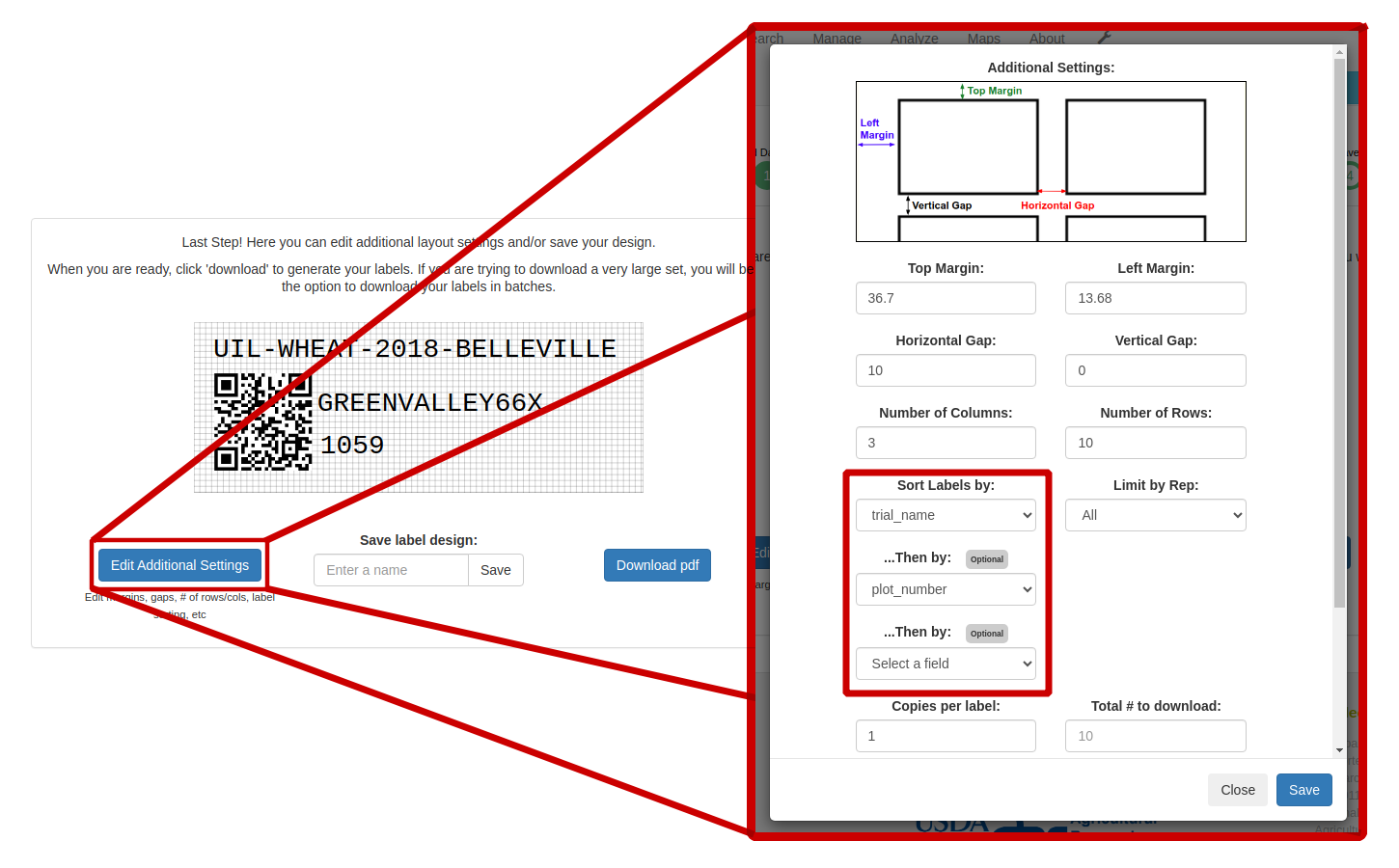
T3 now has the ability to generate plot-level labels for more than one trial at once. To do this, you'll first need to create a list of the trials you want to include (you can quickly make a new list in the Search Wizard). Then, from the data source step of the label designer, select Lists as the data type, your list of trials as the data source, and choose the option to create a label for every plot. Once you're done formatting the label, the output will be a PDF containing a label for each individual plot in all of the trials from your list.
Also, to make it easier to apply the labels once they've been printed, we've expanded the label sorting options. Before the labels are printed, you can sort the individual labels by up to three different properties of the data associated with the labels. For example, if you're printing plot-level labels for multiple trials, you can sort first by trial name and then by plot number. To set the sorting options, click the Edit Additional Settings button and choose the properties to sort by.
T3 Collaborations
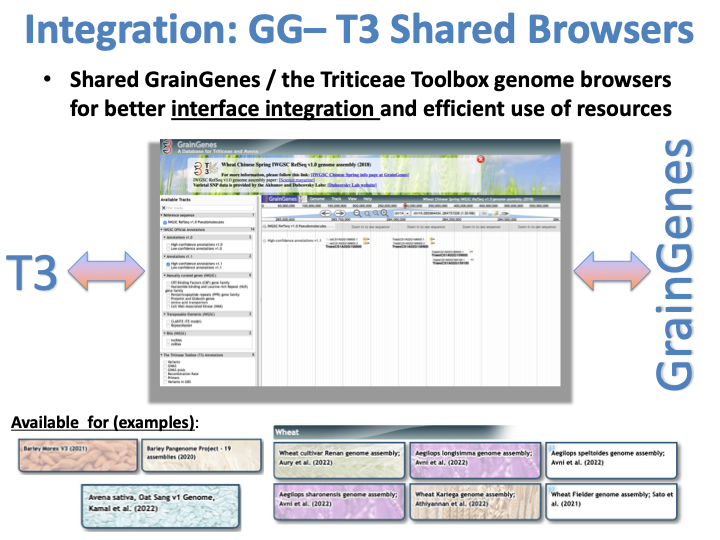
GrainGenes-T3 Shared Genome Browsers
The missions of T3 and GrainGenes are complementary. While the former deals with raw data and tools to help researchers to analyze their data, GrainGenes stores peer-reviewed, processed data for long-term sustainability. In a show of close collaboration between two USDA-ARS databases, GrainGenes and The Triticeae Toolbox (T3) decided in 2019 to maintain shared genome browsers starting with IWGSC Chinese Spring wheat and Morex barley reference genomes. Since then, new shared genome browsers were created and housed at GrainGenes and has been populated by both teams. Users can reach these genome browsers from both databases and link out from browsers to other GrainGenes and T3 pages in a facilitated manner, enabling them to reach a wide range of information in both databases in a way that was not possible before. T3 and GrainGenes will continue their collaboration through joint weekly calls and by looking for ways to share and link data between two sites. If you have ideas for useful links between T3 and GrainGenes, let us know!
Feel free to test out any of these new features and give us any feedback! The quickest way to get in touch with us is the Contact Us button at the top of any page on the T3 websites.
- The Triticeae Toolbox
