Hello,
This newsletter features some updates made to The Triticeae Toolbox (T3) over the last few months, including:
- Improvements for downloading genotype data of selected data from the Search Wizard, retrieving the original VCF files of genotype projects uploaded to T3, and downloading imputed genotype data of select wheat projects.
- A new VCF submission form for submitting genotype data to be included in T3.
- Changes made to the trial and observation uploads to simplify the upload templates.
- A new breeding program summary page that breeders can use to track the progress of their data submitted to T3.
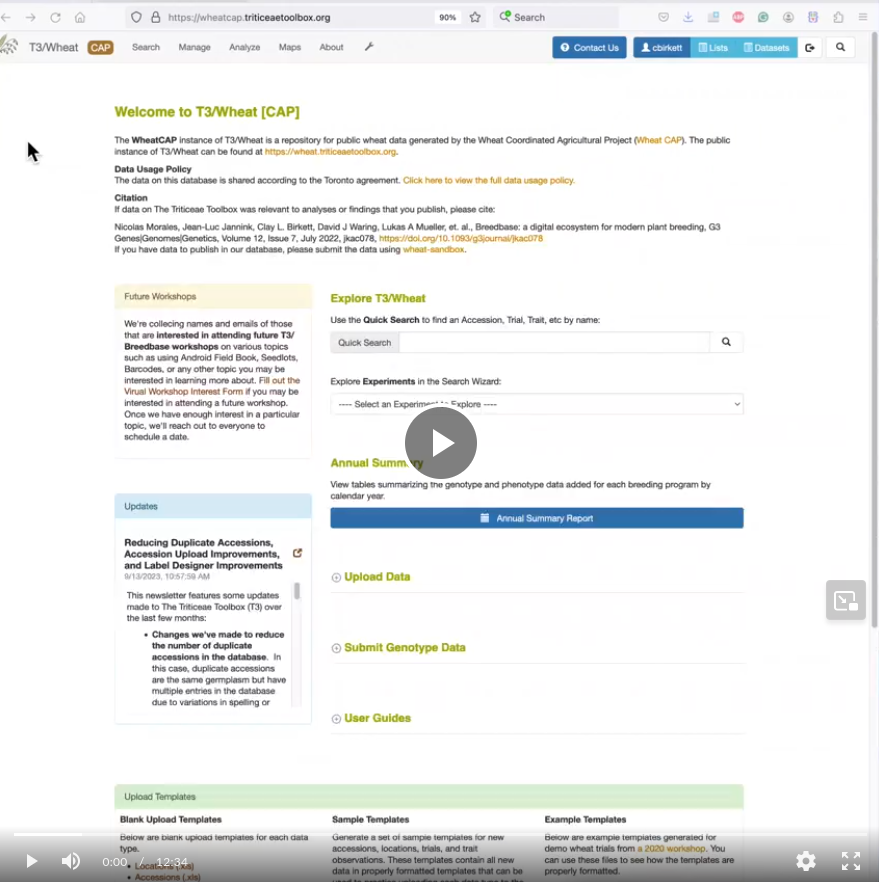
Genotype Data Download
We've made some changes to the Search Wizard to indicate if the selected genotype data will take too long to download. The relative amount of time for a genotype download depends on the number of accessions selected and the number of markers in the selected genotype protocol. If the total number of data points exceeds 1 million, you'll see a warning message that the selected data would likely take a long time to download. In this case, it may be better to download the original archived VCF file of the genotyping project.
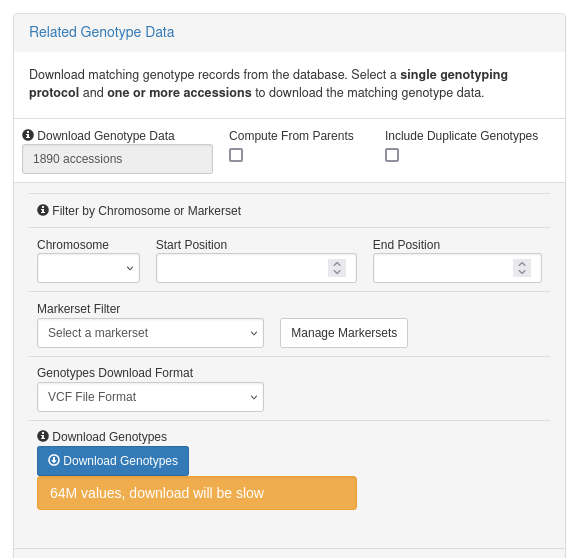
In order to make it easier to download entire datasets, we've added a feature that allows users to download the original archived VCF files of genotyping projects. These are the the original VCF files that were uploaded to T3 when the data was first added to the database. Downloading the original files is generally faster because the VCF files are already stored on the server and don't need to be generated for each request (like the "Related Genotype Data" from the Search Wizard). However, you'll get the entire VCF file and any subsetting or combining of data will have to be done yourself.
Archived VCF files are available from the Search Wizard (under the Archived Genotype Data section) by selecting either a Genotyping Protocol or Project in the Wizard. Downloads are available on a project level and all of the available downloads are listed based on your selected Gentoyping Project and/or Protocol (a protocol may contain multiple projects).

The downloads are also available from a Genotyping Project detail page (accessible from the Search > Genotyping Projects page), under the Archived Files section.
The T3/Wheat (and WheatCAP) sites now have some imputed genotype data available. These selected projects have been imputed using a Practical Haplotype Graph (PHG) constructed using whole-exome capture sequencing data from 472 accessions with 2.89M markers. When a genotype project that has been imputed is selected in the Search Wizard, the imputed data is available under the Imputed Genotype Data section. This will allow you to download a gzipped VCF file of the entire imputed dataset for the selected project.

More information about downloading genotype data from T3 can be found in the Genotype Download using Search Wizard documentation as well as this video tutorial.
Genotype Data Submission Form
In order to streamline the process of submitting genotype data to T3, we've created a new Genotype Data Submission Form. This form is available from the homepage of each of the sites, under the Submit Genotype Data header. This form prompts the user to fill in all of the related metadata about the genotype data (such as the protocol information, description of the sample population, etc). Once the user fills out the form, they'll be directed to a cloud service file submission page where they can upload their VCF file (up to 50 GB in size) for us to access. Once we have the file and the metadata, we can start the process of adding the data to the database.
Upload Template Changes
We've made a minor change to the upload template for submitting phenotype observations. Previously, the first column (which generally has plot names) had to be observationunit_name. We've made it more flexible and easier to copy and paste columns from downloaded files from the website and the first column header can now be either observationunit_name, plot_name, subplot_name, plant_name, observationUnitName, plotName, subplotName, or plantName.
We've also made the trial upload template more flexible. Instead of requiring all of the possible columns in a specific order, only the required columns need to be in the template and they can be in any order. Any optional columns can be added to the template as needed.
In addition, we've added a new optional column to the trial upload template. The entry_number column can be added when you want to assign entry numbers to accessions in a trial. These entry numbers are stored at the trial level (so different trials can have different entry numbers for their entries) and are included in trial-level downloads. We're working on making them available to the barcode label designer so they can be included on printed barcodes.
Breeding Program Summary Page
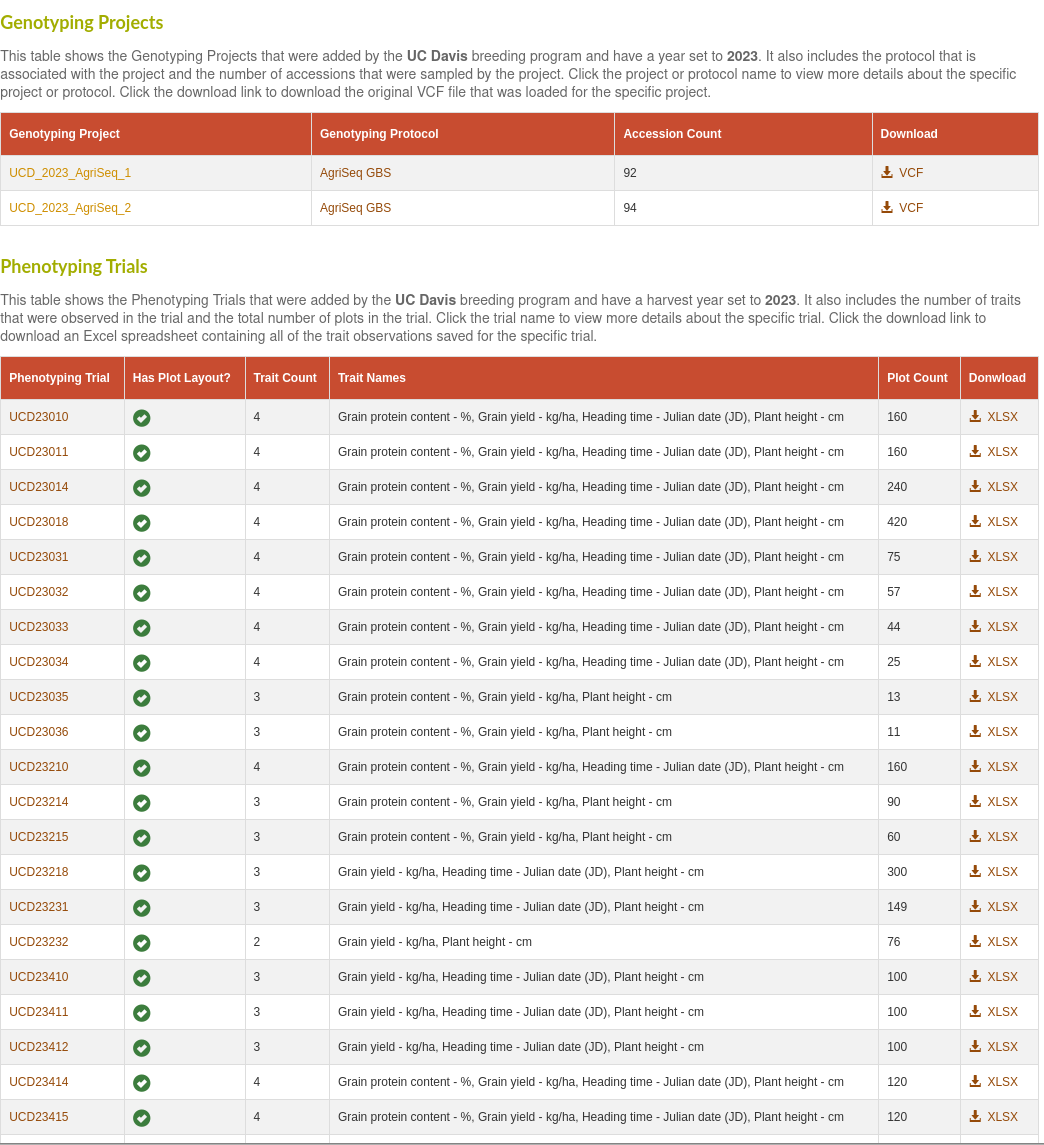
Lastly, we've added a new annual summary report page to track a breeding program's phenotype and genotype data submissions to the database. This will summarize the number of phenotype trials and genotype projects added to the database by a breeding program for each calendar year. It will also indicate if there are any trials that don't yet have observations added.
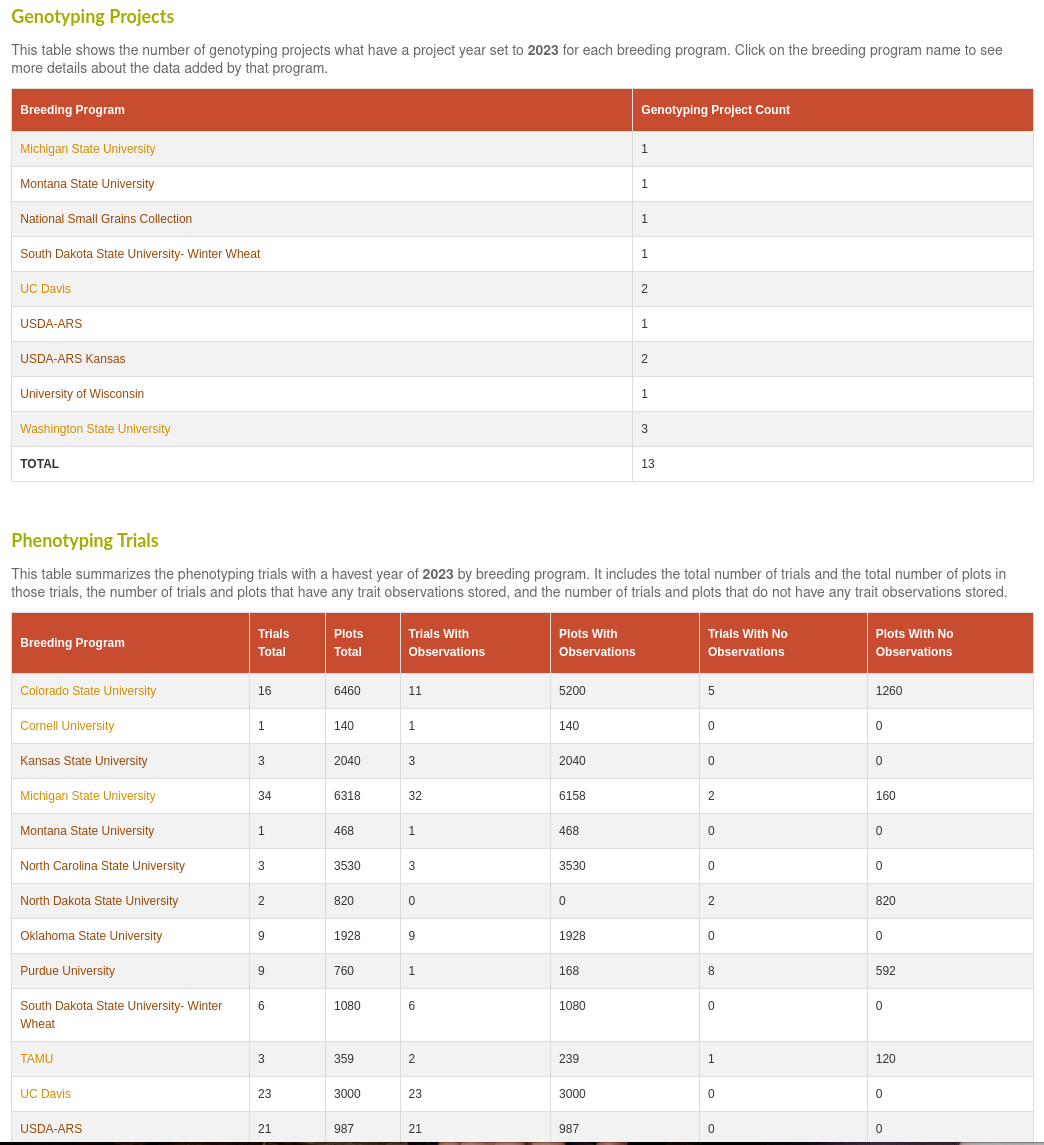
If you select a particular breeding program, you will get a list of the individual genotype projects and phenotype trials, with links to directly download the data associated with those individual projects. The genotype table will include the number of accessions that were sampled by each genotype projects. The phenotype table will include a check for each trial that has a defined plot layout, the number of plots, the number of traits, and a list of the trait names observed for each trial.
As always, feel free to test out any of these new features and give us any feedback! The quickest way to get in touch with us is the Contact Us button at the top of any page on the T3 websites.
- The Triticeae Toolbox